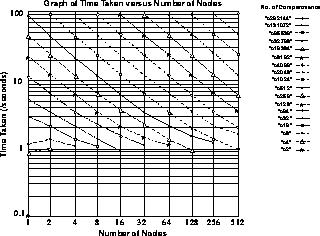
Figure 7.14: Results from Running a GENESIS Simulation of a Passive Cable Model Composed of Varying Numbers of Compartments Distributed Across Varying Numbers of Nodes on the Intel Touchstone Delta Parallel Supercomputer




The postmaster element is a self-contained object within the GENESIS neural simulator. Like the objects in a true object-oriented language it is an entity composed of public (externally available) data, private (internal) data, and functions to operate on the object. Like other GENESIS elements, it is a black box that can be used to construct a neural simulation. For the present, modellers wishing to produce large distributed simulations that are able to take full advantage of the power inherent in our current parallel computers must specify how to distribute the different parts of their simulation over the available hardware computing nodes. While this may be an annoying necessity to certain neural modellers, it must be remembered that this technique allows the construction of simulations of such a size and computational complexity that they can be modelled on no other existing computer platforms (e.g., traditional serial supercomputers). Bearing this in mind, the requirement of explicitly stating how to distribute the model is a small price to pay for the power gained. This explicit method also brings with it certain advantages. Firstly, because the modeller is familiar with the computational demands of the different parts of his model, he is able to accurately balance the computational load over the available parallel hardware. This makes for far more efficient load balancing and scaling behavior than would be possible with an automatic decomposition scheme that has to balance the very different needs of both single-cell modellers and network-scale modellers. It is also less efficient to carry out automatic decomposition because the various GENESIS elements comprising a complete neural simulation have widely varying computational demands. As with other GENESIS elements, the postmaster acts as a self-contained black box which usually communicates information about its changing state via messages to other elements. It can both send messages to, and receive messages from, other GENESIS elements which may exist either on this particular hardware node or on others in the network. As a result of this, it is an element that ties together and coordinates the disparate parts of a model running on separate hardware nodes of the parallel computer. The actual messages transferred depend on the type of element with which it communicates. The postmaster element neither knows nor cares whether the quantity it is communicating is a membrane potential, a channel conductance (simple current flow), or the concentration of any substance from ions to complex neurotransmitters. The postmaster is a two-faced element. Its first face is that of a normal GENESIS element which it presents to the rest of the simulation. This first aspect of the postmaster is able to pass GENESIS messages between elements and allows use of the GENESIS Script Language to query its state. The second aspect of the postmaster is aware of the parallel nature of the underlying hardware and can use the operating system primitives for communicating information between separate physical nodes. In summary, the postmaster element is a conduit along which information can flow between nodes. It allows the modeller to tie together the disparate aspects of the simulation into a coherent whole.
To show how this mechanism works in practice, performance measurements are presented in Figure 7.14, which are taken on the Intel Touchstone Delta parallel supercomputer based at Caltech. These figures show both scaling of performance, where more nodes allow the model to complete in a shorter timescale, and the construction of models so large that it has been impossible to model them before now.

Figure 7.14: Results from Running a GENESIS Simulation of a Passive Cable
Model Composed of Varying Numbers of Compartments Distributed Across
Varying Numbers of Nodes on the Intel Touchstone Delta Parallel
Supercomputer
The previous record for the most complex GENESIS model produced on a
traditional serial computer is approximately 80,000 elements, the
Purkinje Cell model produced at Caltech by
Dr. de Schutter [Schutter:91a;93a].
Using the postmaster
element on a parallel supercomputer (the 512-node Intel Touchstone
Delta), this limit has now been pushed to over two million GENESIS
elements (actually 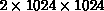 ). As can be seen, this now
allows for the construction of far more complex and realistic models
than was previously possible. The present price to be paid for this
freedom is the decision to make the modeller explicitly distribute his
simulation over the available hardware. This was a design decision that
has allowed far greater efficiency of load balancing than would be
possible using an automatic distribution technique as the illustrated
scaling graphs confirm. This technique also has the advantage of
leaving the basic GENESIS script interface unaltered and is applicable
to a wide variety of parallel hardware. As a result of the requirement
to retain compatibility with the existing serial GENESIS implementation,
another benefit has become obvious. By changing only the network layer
of the postmaster element, it is possible to produce a version of the
postmaster that can use traditional serial machines distributed across
the Internet to produce a distributed model, which ties together
existing supercomputers based anywhere on the network. The potential
size of model that can be constructed in this manner is staggering,
although the reality of network communication delays will limit its area
of application to compute-limited tasks (cf. communication-bound
simulations).
). As can be seen, this now
allows for the construction of far more complex and realistic models
than was previously possible. The present price to be paid for this
freedom is the decision to make the modeller explicitly distribute his
simulation over the available hardware. This was a design decision that
has allowed far greater efficiency of load balancing than would be
possible using an automatic distribution technique as the illustrated
scaling graphs confirm. This technique also has the advantage of
leaving the basic GENESIS script interface unaltered and is applicable
to a wide variety of parallel hardware. As a result of the requirement
to retain compatibility with the existing serial GENESIS implementation,
another benefit has become obvious. By changing only the network layer
of the postmaster element, it is possible to produce a version of the
postmaster that can use traditional serial machines distributed across
the Internet to produce a distributed model, which ties together
existing supercomputers based anywhere on the network. The potential
size of model that can be constructed in this manner is staggering,
although the reality of network communication delays will limit its area
of application to compute-limited tasks (cf. communication-bound
simulations).
The assessment of the usefulness of a parallel neural modelling platform can be demonstrated by two extremes of neural modelling applications:
The individual subcompartments which make up a neuron's dendritic tree are active simultaneously, and each of these compartments may be studded with several independently functioning ionic membrane channels. This is illustrated in Figure 7.15.
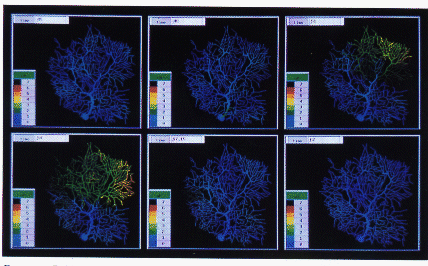
Figure 7.15: Cerebella Purkinle cell model in GENESIS.
Our most detailed single-cell model to date.
A detailed network simulation may be composed of many thousands of individual nerve cells from a smaller number of biological cell types.
What does a parallel computer have to offer the single-cell modeller?
Most of the work on the parallel GENESIS at Caltech to date has been in the field of single-cell modelling. Several distinct ways of using the system have been developed allowing a variety of approaches by the single-cell modeller.
Each individual node on the parallel computer runs a separate, complete, single-cell model (task farming). This facility can be used to examine the sensitivity of the cell's performance to a wide range of physiological states including testing the effect of parameters which are at present difficult to measure experimentally [Bhalla:93a]. A selection of these appear below:
Like the experimentalist, the neural modeller can block or poison different ion channel subsets, with the added advantage that it is possible to block both channels where no chemical blocking agent currently exists and also to have 100% channel-blocking specificity (e.g., work conducted by D. Jaeger, Caltech [Jaeger:93a]).
An experiment at present difficult or impossible to perform in any physiological setup can easily be tested on the computer model system. For example, the independence of stimulation site on Purkinje Cell response (Work performed by E. de Schutter, Caltech [Schutter:91a;93a]).
The prediction of ionic channel distribution over different parts of a cell's dendritic arbor by observing the effect of changed distribution on the model cell's electrophysiological properties, and relating this to the experimentally measured behavior of the real neuron [Bhalla:93a]. Experimental confirmation of channel distribution predicted by the manipulation of such computer models seems likely to appear in the near future due to advances in monoclonal antibody techniques for different channel subsets.
Predicting the effect of changing membrane properties which are impossible to measure experimentally-for example, in the distal dendritic arbor or in ``spines'' (e.g., E. de Schutter, Caltech [Bhalla:93a], [Schutter:91a]).
The first example above is of modelling following and confirming physiology experiments. The latter examples are uses of neural modelling to predict future experimental findings. Although rarely used to date (because of computer limits on the model's level of biological realism), this synergistic use of neural modelling in predicting experimental results and suggesting new experiments appears to offer substantial benefits to the neurobiology community at large. Neural modelling on parallel computers, such as the Intel Touchstone Delta, is allowing modelling to adopt these new closer links to experimental work, thereby closing the dichotomy between experimenters and modellers. In the past, this dichotomy has caused several experimentalists to question the relevance of funding modelling work. Hopefully, this attitude will change as more results of synergy between modelling experiments and physiology experiments become widely known.
The system allows the construction of larger and more detailed cell models than is possible on a traditional serial computer. The level of detail included in models to date has been limited either by the memory size constraints of the computer used, or by the computational time requirements of the model [Bhalla:92a]. A distributed model of a single cell on a parallel computer alleviates both of these constraints simultaneously. This allows the construction of larger cell models than have been previously possible but which nevertheless run in acceptable time frames.
What does a parallel computer have to offer the neural network modeller?
Much of the work to date has been on task farming (as described above), whereby each node runs its own copy of a cell. This is less useful to the network modeller but still allows detailed statistical information to be built up about network and population behavior. A more interesting way of using the parallel machine for network modelling is the distributed model scheme mentioned above. This allows networks to be both larger and to run more quickly than their counterparts on equivalent serial machines. A promising project in this category, although still in its very early stages, is the construction by Upinder Bhalla at Caltech of a detailed model of the rat olfactory bulb [Bhalla:93a]. This incorporates detailed cellular elements, which are rare in network class models. Such a network model makes far greater communication demands of the internode communications mechanism on the parallel computer than a distributed single-cell model. Initially an expanded version of the postmaster element [Speight:92a;92b] which was used for distributed single-cell models will be employed, but this may change as the different demands of a large network model become apparent.
All of the work on the Parallel GENESIS project was carried out in the laboratory of Professor James Bower at the California Institute of Technology.