- 3.1 Operation of the Web Forager
- 3.2 Parallelizing the Forager
- 3.3 Foraging Quietly
- 3.4 Web Forager Implementation Notes
- 4. Software Submissions, Cataloging, and Review
- 5. User Profiles
- 6. HPCC Thesaurus and Roadmaps
- 7. Future work
- 7.2 Partially Automated Cross-referencing and Classification
- 7.3 Relevance Feedback for Searching
- References
1. Background and History
The National HPCC Software Exchange (NHSE) is funded by NASA as part of the High Performance Computing and Communications (HPCC) program, established in 1991. NASA's primary role in the HPCC program is to lead the development of applications software and algorithms for scalable parallel computing systems. NASA is also responsible for fostering software sharing and reuse across federal HPCC programs. The NHSE is under development by the Center for Research on Parallel Computation (CRPC).The purpose of the NHSE is to provide access to all software and software-related resources produced by the HPCC Program. Access is provided in a manner that promotes and facilitates reuse and technology transfer policies and processes established by HPCC program agencies. The NHSE provides a uniform interface to a distributed collection of networked repositories, which for administrative and logistical reasons are maintained separately. By using semi-automated submission and indexing mechanisms, the NHSE imposes only minimal delay between the production and distribution of software resources.
Although the different disciplines will maintain their own software repositories, users need not access each of these repositories separately. Rather, the NHSE provides a uniform interface to a virtual HPCC software repository that will be built on top of the distributed set of discipline-oriented repositories. The interface assists the user in locating relevant resources and in retrieving these resources. A combined browse/search interface allows the user to explore the various HPCC areas and to become familiar with the available resources. A long-term goal of the NHSE is to provide users with domain-specific expert help in locating and understanding relevant resources.
The target audiences for the NHSE include HPCC application scientists, computer scientists, users of government supercomputer centers, and industrial users. The expected benefits from the NHSE include:
- Faster development of high quality software so that scientists can spend less time writing and debugging programs and more time on research problems.
- Reduction of duplication of software development effort by sharing of software modules.
- Reduction of time and effort spent in locating relevant software and information through the use of appropriate indexing and search mechanisms and domain-specific expert help systems.
- Reduction of the time scientists spend dealing with information overload through the use of filters and automatic search mechanisms.
- Systems software and software tools. This category includes parallel processing tools such as parallel compilers, message-passing communication subsystems, and parallel monitors and debuggers.
- Data analysis and visualization tools.
- High-quality transportable building blocks for accomplishing common computational and communication tasks. These building blocks are meant to be used by Grand Challenge teams and other researchers in implementing programs to solve computational problems. Use of high-quality transportable components will speed implementation and will increase the reliability of computed results.
- Research codes that have been developed to solve difficult computational problems. Many of these codes will have been developed to solve specific problems and thus will not be reusable as is. Rather, they will serve as proofs of concept and as models for developing general-purpose reusable software for solving broader classes of problems.
2. Manual Information Collection
The current NHSE information collection has been constructed by manually generating and maintaining sets of HTML pages at the different CRPC sites. These pages contain information about HPCC, as well as pointers to external URLs relevant to HPCC. The NHSE home page, as well as a searchable index to the distributed NHSE information collection, is maintained at the University of Tennessee site. Other sites with large collections of NHSE URLs include NPAC at Syracuse University, Argonne National Laboratory, and Rice University.The URLs were discovered by contacting groups and individuals active in HPCC, by searching for HPCC-related information on the Web, and by soliciting user contributions. The URLs have been organized into categories and sub-categories to facilitate browsing. One of the major categories is HPCC software and enabling technologies, which includes the NHSE software catalog. This catalog contains close to 300 pieces of software in the categories of benchmark programs, data analysis and visualization, numerical programs and routines, parallel processing tools, and scientific and engineering applications.
Over 1000 URLs have been collected, and more are added every day. Many of these top-level URLs are lists that point to other relevant URLs. By going just two levels down from the NHSE home page, the user may access a total of over 15,000 HTML pages relevant to HPCC.
The Harvest system [3] provides the search interface to the NHSE collection. A Harvest gatherer running at the University of Tennessee retrieves all URLs pointed to by the NHSE, plus one additional level -- in other words, three levels of a breadth-first search tree rooted at the NHSE home page. A Harvest broker indexes the files using WAIS and provides a query interface for keyword searching. Other sites will soon be running Harvest gatherers as well, so that deeper and more comprehensive indexing may be carried out. Because gatherers stream summary information to brokers, rather than sending files individually, such a distributed setup permits efficient use of network bandwidth. As the NHSE broker becomes more heavily used in the future, it is likely to become overloaded. To deal with this problem, Harvest provides for broker replication. We will monitor the load on the NHSE broker and replicate it as needed.
3. Automatic Information Collection
As part of the research in enabling technologies for the NHSE, Argonne is building a toolkit for exploring advanced Web resource management technologies. The toolkit will support hunting and gathering Web pages (http, ftp, gopher), compression, indexing, transaction monitoring, parallel search and a rich language environment for developing agents. The toolkit includes a modular Web forager, a parallel Web indexing engine, and autonomous search agents.The modular programmable Web forager is designed to efficiently cache Web pages on a local server, based on programmable starting locations, keywords, file types, and other search criteria. The Web forager runs in parallel to allow high-performance gathering of Web pages. Its modular design allows it to be easily modified.
3.1 Operation of the Web Forager
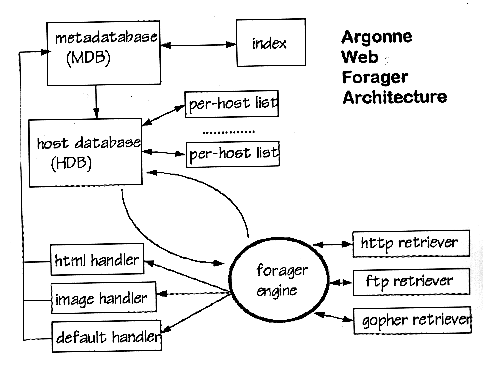
A schematic of the Web forager is shown in Figure 2.- Initialize the URL pool
- While there are pages in the pool
- Remove a URL from the pool
- Download that page from the Web
- Parse the page, generating a new list of URLs. Add these URLs to the pool.
- Index the page
- If caching, save the page locally
We maintain a database of information about the URLs that have been visited (the meta-database). The information is kept on a per-URL basis and includes
- Date page retrieved
- Last modification date of document on server
- Document type
- Retrieval frequency
- Expiration date
- Last time host was contacted
- Preferred interval between accesses to host
- List of URLs to be searched
- Initialize the URL pool
- While there are URLs to be searched
- Pick a host to search
- Remove a URL from that host's URL pool in the host database
- Download that page from the Web
- Parse the page, generating a new list of URLs.
- Update the meta-database with each new URL
- Add each URL to its host's URL pool in the host database
- Index the page
- If caching, save the page locally
- Update the meta-database entry for the searched URL
3.2 Parallelizing the Forager
One of the goals of the forager project is to complete a five million URL forage in a week. Such a run introduces two obstacles: network latencies and the necessity of handling large databases.We have seen from our experiments with a sequential forager that most of the time spent in the forager is spent in waiting for the wide area network -- over eighty percent of a recent run. Because in a large run there will be a large number of servers from which to retrieve pages, we can very efficiently utilize multiple foragers to overlap useful computation and the time spent waiting on the network. The forager processes are very lightweight, carrying little state information. As a result we can usefully run multiple foragers on each node in the computation.
The size of the databases required for a five million URL forage introduce some practical problems. We want to keep the databases in memory as much as possible, as the operation of the forager requires both frequent lookups into the databases (for URL pool and host information) and frequent updates to the databases (new entries to the URL pool, updates to the metadatabase). We anticipate a metadatabase size of over 1.5 gigabytes (not including the URL pools). The parallel forager distributes the meta- and hostdatabases across several computers, reducing the size of the individual databases to the point that much of the database will be cachable in memory.
3.3 Foraging Quietly
We have taken great care in the design of the forager to make it a good network citizen. Other web robots have caused problems by making rapid-fire requests to a single server, repeatedly requesting the same file from a server, or by making large numbers of meaningless requests (for instance, making deep traversals in virtual trees or by invoking CGI scripts with side effects). These problems led to the development of the Standard for Robot Exclusion [9]. The NHSE web forager is compliant with this standard.We also adhere to the philosophy that the forager should be no more intrusive than a human browsing the web. We implement this by allowing the forager to access any given host for only a short period of time (defaulting to thirty seconds) before moving on to the next host. Once a search has been running for a while, there are enough hosts to be searched that the search frequency for any given host is not high. In addition, host-specific requirements (search frequency, interval between successive request, and so on) can be specified on a per-host basis in the case that the default policy is not appropriate.
3.4 Web Forager Implementation Notes
The Web forager is written in Perl 5 [14] for modularity and rapid prototyping. Communication is handled by using Nexus [6], which provides remote procedure call semantics, threading support, remote reference support, and clean integration with Perl 5. Database support is via the DB_File interface module [11] Berkeley DB library [13], which provides disk-based databases with efficient in-memory caching. The modular design of the forager permits the developer to plug in per-document-type handler modules, as well as per-protocol retrieval modules.3.5 Parallel Web Indexing Engine
Argonne is also developing a parallel extension of the Glimpse (University of Arizona) indexing system [10] for rapidly indexing web pages (*.html and other file types) on parallel systems and for providing rapid regular expression based parallel searches of Web page caches, such as those generated by our Web forager. We are also developing extensions to the query system specifically allowing us to locate "software" in the midst of other Web information, thus supporting the ability to search for data that contains software (source files, binaries, tar files, makefiles etc.) across the Web. This Web indexing engine should in principle be scalable to millions of URLs. A five million URL test run is planned for the near future.The indexing structure distributes indices across multiple index nodes, allowing lookups to be carried out in parallel. Content pages may be discarded after indexing, because the Glimpse indexing mechanism can be used to identify URLs to be retrieved for further filtering. A Web form-based search interface is provided.
The most recent testbed search database, collected January 25 - 29 1995, contains 52032 URLs, including 37700 HTML pages, from 13000 sites. To generate this database, the Web forager was started from the Argonne Mathematics and Computer Science Division home page.
4. Software Submission, Cataloging, and Review
Contributors submit software to the NHSE by filling out the NHSE Software Submission Form, accessible from the NHSE homepage. The form explains the submission and review process, including the authentication procedures, and gives an example of a completed submission form. The form asks the user to fill in values for several attributes, some required and some optional. Contributors submit software for consideration at a particular review level. Currently three levels of software are recognized in the NHSE:- Unreviewed. The submission is not reviewed by the NHSE for conformance with software guidelines.
- Partially reviewed. The submission undergoes a partial NHSE review to verify conformance with the scope, completeness, documentation, and construction guidelines. These particular guidelines are those that can be verified through a visual inspection of the submission.
- Reviewed.The submission undergoes a complete NHSE review to verify conformance with all the software guidelines. This classification requires peer-review testing of the submitted software. This level may be further refined into additional levels in the future.
- Scope. Software submitted to the NHSE should provide a new capability in numerical or high-performance computation or in support of those disciplines.
- Completeness. Submissions must include all routines and drivers necessary for users to run the software. Test problem sets and corresponding drivers must be included if the software is to undergo peer-review testing for the Reviewed level. Source code for widely available software used by the submission, blas and lapack for example, need not be included as part of the submission.
- Documentation. The software contains complete and understandable documentation on its use.
- Construction. Submissions must adhere to good mathematical software programming practice and, where feasible, to language standards. Software should be constructed in a modular fashion to facilitate reusability. The use of language checking tools, such as pfort or ftnchek, is recommended.
Software submitted for full review is reviewed according to the following criteria:
- Documentation. The software contains complete, understandable, correct documentation on its use.
- Correctness. The software is relatively bug-free and works as advertised on all provided data sets and on data sets provided by the reviewer according to the documentation..
- Soundness. The methods employed by the software are sound for solving the problem it is designed for, as described in the documentation.
- Usability. The software has an understandable user interface and is easy to use by a typical NHSE user.
- Efficiency. The software runs sufficiently fast to make it an effective tool.
Once the software has been reviewed, one of two things happens. If it is not accepted, the author will be so informed and anonymous copies of the reviews will be provided. The author may then choose to address the reviewers' comments and resubmit revised software. If the software is accepted, the author will be shown a review abstract summarizing the reviewer comments. This abstract will be available to anyone who accesses the software through the NHSE. If the author finds the abstract unacceptable, he or she may withdraw the software and resubmit it for review at a later date.
After they have been processed, software submissions are placed into the NHSE software catalog. The cataloging process is carried out jointly by the authors and the NHSE librarian, with the authors providing the title and abstract fields, and the NHSE librarian categorizing each entry and assigning thesaurus keywords. The NHSE software catalog is available in the following formats:
- An HTML version that may be browsed by category.
- A searchable version that allows the user to search separately by different attributes or to do a full-text search on the catalog records. A link to an on-line copy of the HPCC thesaurus is provided so that users may select controlled vocabulary terms for searching. The current interface requires users to cut and paste thesaurus terms into the search form. We plan to develop a hypertext version of the thesaurus that will statically link thesaurus terms to scope and definition notes and to related terms as well as dynamically link thesaurus terms to indexed catalog entries.
5. User Profiles
A user may submit a user profile by filling out the NHSE User Profile Submission Form, accessible from the NHSE homepage. This form asks the user about his or her background, interests, and software and information needs. Although name and email address are requested so that a reply can be made, a user profile is kept confidential unless the user gives permission to publish it.The purpose of collecting user profiles is two-fold:
- To serve NHSE users by providing customized responses to requests for information.
- To collect a sample database of profiles to use for comparative testing of different search strategies.
The user profile database will also provide a test query set for evaluating the recall and precision of the following search strategies:
- natural language processing (NLP) [12] alone
- Latent Semantic Indexing (LSI) [4]alone
- LSI with NLP noun phrase extraction as a preprocessing step
- using the HPCC thesaurus for both manual indexing and searching with boolean searches
- using the HPCC thesaurus as a searching thesaurus only for boolean searches
- NLP assisted by thesaurus scope notes and definitions
- LSI assisted by thesaurus scope notes and definitions
6. HPCC Thesaurus and Roadmaps
6.1 HPCC Thesaurus
An HPCC thesaurus is currently under development as part of the NHSE development effort. This thesaurus is being developed according to the ISO 2788 thesaurus standards [8] using a faceted construction technique [2] for the core area of mathematical software. Other sources of vocabulary for the core areas are the current NHSE contents, the HPCC glossaries described below, and the book Parallel Computing Works [7].The HPCC thesaurus is intended to be used directly by NHSE users rather than by expert search intermediaries. Therefore, the use of complex devices will be minimized, and extensive scope notes and definitions of thesaurus terms will be provided. Thesaurus terms will be assigned manually to NHSE Software Catalog entries by the NHSE librarian as part of the submission process. Although the larger body of NHSE informational HTML pages will not be indexed manually, the thesaurus will still be useful for searching this collection, as it will provide an overview of the field and will supply candidate terms for searching.
6.2 Glossaries and Other Roadmaps
In view of the positive reception of our HTML glossary on HPCC terminology at Supercomputing 94, we have developed new glossaries on other subjects relevant to the NHSE. In particular we are building general glossaries on HPCC applications areas, HPCC software technologies, and specialist glossaries of terms and keywords in High Performance Fortran.We have given a lot of thought to the concept of a glossary and on how it relates to other ways of packaging information on the WWW. The traditional glossary is a way of explaining ``jargon'' in a textbook and is distinguished from other alphabetically sorted lists of definitions such as dictionaries, encyclopedias and thesauri not only by the ``granularity'' of the information entities but also by the way entries are cross referenced. Conventionally, a glossary lists definitions of keywords, acronyms or key phrases in a fairly informal prose style, with cross references (if any) indicated by italic or some other form of printing emphasis. Our HTML glossaries are written in a similar style, with italic font used to indicate internal cross references and bold font for external references. The main distinguishing feature is that the HTML form allows the references to be URLs, either internal ones in the hash format, or full http form external references to other information entities on the Web.
We view the glossary form as a good form to encapsulate domain level expertise on a particular subject, whether that be a broad subject such as HPCC or a narrower subject such as the particular HPCC terminology associated with High Performance Fortran for example.
One disadvantage of the glossary form is its non-scalability. To be successful a glossary has to be written from a particular point of view and must have a consistent philosophy. We believe this is relatively straightforward when the subject covered is such that an individual or a small editorial team can carry out the entire review process. This becomes harder for multi-disciplinary subjects such as ``HPCC Applications''. For this reason we are currently working on ways of linking the glossary and thesaurus concepts into a consistent hierarchy of information systems.
We envisage the following conceptual hierarchy of information granules:
- Specific real items of software for applications, libraries, tools, languages, or environments.
- Hypertext ``Encyclopedia articles'' that are written from a particular perspective, and are essentially review articles or technical notes.
- Glossaries that are essentially ``hypertext expert systems'' or review articles with a lower granularity than encyclopedia articles.
- Thesauri, which are generalized cross referencing mechanisms, coupled to search engines.
The HPCC glossary was first initiated as an HTML document with hand written entries with embedded HTML tags. This approach is both cumbersome and error prone. To partially solve these difficulties we have developed a scripting tool that parses entries and flags incorrectly formatted entries and makes checklists of internal, external, and duplicated entries. However, as the HPCC glossary alone has grown to over 700 definitions, this approach is no longer scalable, since it becomes increasingly difficult for a human being to keep track of the possible cross references when writing a new entry. We have developed textual analysis tools to aid the entry writer in cross referencing a new entry. We are using hypertext technology for this in the form of client data forms that invoke word stemming and other analysis programs on the server side. The word stemming process is required so that duplicate cross references are not necessary for an entry that might be ``cache'', ``caches'', ``cached'', or ``caching'' for example. We have built tools that can parse glossary entries from manually edited hypertext, as well as from HTML forms. The resulting entries may be input directly into the on-line glossary system or submitted to an editorial review board.
We have developed a general roadmap navigation package to HPCC related sites and activities with an attempt to link applications activities to software and other technical information stored in the NHSE. We are currently integrating this with the glossary and thesaurus systems.
The NHSE roadmaps and glossaries are all accessible from the "roadmap" link on the NHSE home page.
7. Future Work
7.1 Heuristic Information Collection
In our manual information collection, we evaluate contributed HTML pages and only add those to our collection that are within the scope of high performance computing. When browsing the Web manually to collect information, we selectively follow those links that appear most relevant. As currently implemented, our Web forager, when started from a particular location, follows all links with equal likelihood. We plan to extend the forager with filtering mechanisms, based on keywords, file types, and semantic analysis. These heuristics will enable selective retrieval of the most relevant information. Semantic analysis using techniques such as LSI [4] will allow automatic selection of documents that are similar, or "close in the concept space", to the manually selected collection. An option of an interface interface with a human operator will allow human feedback to guide the forager's Web traversal.7.2 Partially Automated Cross-referencing and Classification
Classification and cross-referencing tasks take place at several points in the NHSE's information management processes. When a contributor submits a new software package, the NHSE librarian places it into the appropriate category and assigns thesaurus keywords. Newly contributed URLs for Web pages are placed on the appropriate list on the appropriate NHSE page. New HPCC glossary entries are cross-referenced to existing entries and to external material. Now that a fair amount of manual classification has been carried out, we are in a position to begin partially automating the classification process. Using both previously classified material and newly contributed material as inputs, semantic analysis techniques may be used to suggest appropriate classifications for the new material. Preliminary experiments we have carried out using LSI to generate candidate GAMS classifications for mathematical software have shown close agreement with the classifications assigned by human experts. Although LSI and related techniques are not accurate enough to completely automate the classification process, partial automation will allow a much larger amount of material to be processed with the same manpower, and may reveal unexpected linkages that would have been missed by human classifiers.7.3 Relevance Feedback
As with most current Web search services, the current NHSE search interfaces are limited to keyword searching. Because of vocabulary mismatch problems, and because users are unsure what keywords to enter, free-text keyword searching results in poor recall. Recall can be improved by allowing the user to iteratively refine a search and to apply relevance feedback to search results. We plan to implement relevance feedback capabilities for our search interfaces that will allow the user to select one or more items of particular interest for the purpose of augmenting a previous search. Both the NTTC natural language processing [12] and the LSI search engines provide this capability, and we will be experimenting with both of these systems.7.4 Customized Filtering
Automatic filtering and profiling techniques that are purely keyword based have shown poor results, because of vocabulary mismatch problems, but also because the relevance of a particular item is often implicit in its context. We plan to use semantic analysis, augmented by relevance from users, to automate foraging of information to match submitted user profiles. Some previous work in this area is described in [5].References
- 1
- Jean Aitchison and Alan Gilchrist. Thesaurus Construction: A Practical Manual, 2nd ed. Aslib, London, 1987.
- 2
- Ronald F. Boisvert and S. E. Howe and D. K. Kahaner. The Guide to Available Mathematical Software problem classification system, Comm. Stat. - Simul. Comp. 20(4), 1991, pp. 811-842. (GAMS is available on-line at http://gams.nist.gov/).
- 3
- C. Mic Bowman, Peter B. Danzig, Darren R. Hardy, Udi Manber, Michael F. Schwartz, and Duane P. Wessels. Harvest: A Scalable, Customizable Discovery and Access System. Technical Report CU-CS-732-94, Department of Computer Science, University of Colorado, Boulder, August 1994 (revised March 1995). See http://harvest.cs.colorado.edu.
- 4
- S. Deerwester and S. Dumais and G. Furnas and T. Landauer and R. Harshamn. Indexing by Latent Semantic Analysis. Journal of the Americal Society for Information Science 41(6), September 1990, pp. 391-407.
- 5
- Peter W. Foltz and Susan T. Dumais. Personalized Information Delivery: An Analysis of Information-Filtering Methods. Communications of the ACM 35 (12), December 1992, pp. 51-60.
- 6
- Ian Foster, Carl Kesselman, and Steven Tuecke. Nexus: Runtime Support for Task-Parallel Programming Languages. Technical Memo ANL/MCS-TM-205, Argonne National Laboratory, 1995. . http://www.mcs.anl.gov/nexus/paper/index.html.
- 7
- Geoffrey C. Fox, Roy D. Williams, and Paul C. Messina. Parallel Computing Works. Morgan Kaufmann, 1994. (available on-line at http://www.infomall.org/npac/pcw/).
- 8
- International Organization for Standardization. ISO 2788: Guidelines for the establishment and development of monolingual thesauri, 2nd ed. Geneva: ISO, 1986.
- 9
- Martijn Koster. A Standard for Robot Exclusion. http://web.nexor.co.uk/users/mak/doc/robots/norobots.html.
- 10
- Udi Manber, Sun Wu, and Burra Gopal. Glimpse: A tool to search entire file systems. http://glimpse.cs.arizona.edu:1994/.
- 11
- Paul Marquess. DB_File - Perl5 access to Berkeley DB. http://www.mit.edu:8001/perl/DB_File.html.
- 12
- Joe Ross. NTTC Digital Library: A Robust, Replicable Package. INFOTECH '94: DOE Technical Information Meeting, Office of Scientific and Technical Information, October 1994, pp. 71-18.
- 13
- Margo Seltzer and Ozan Yigit. A New Hashing Package for UNIX. USENIX, Winter 1991, Dallas, Texas. (BerkeleyDB is available at ftp://ftp.cs.berkeley.edu/pub/4bsd/db.tar.Z).
- 14
- Larry Wall. Perl 5 Manual. http://www.metronet.com/0/perlinfo/perl5/manual/perl.html.
browne@cs.utk.edu